Note
Go to the end to download the full example code
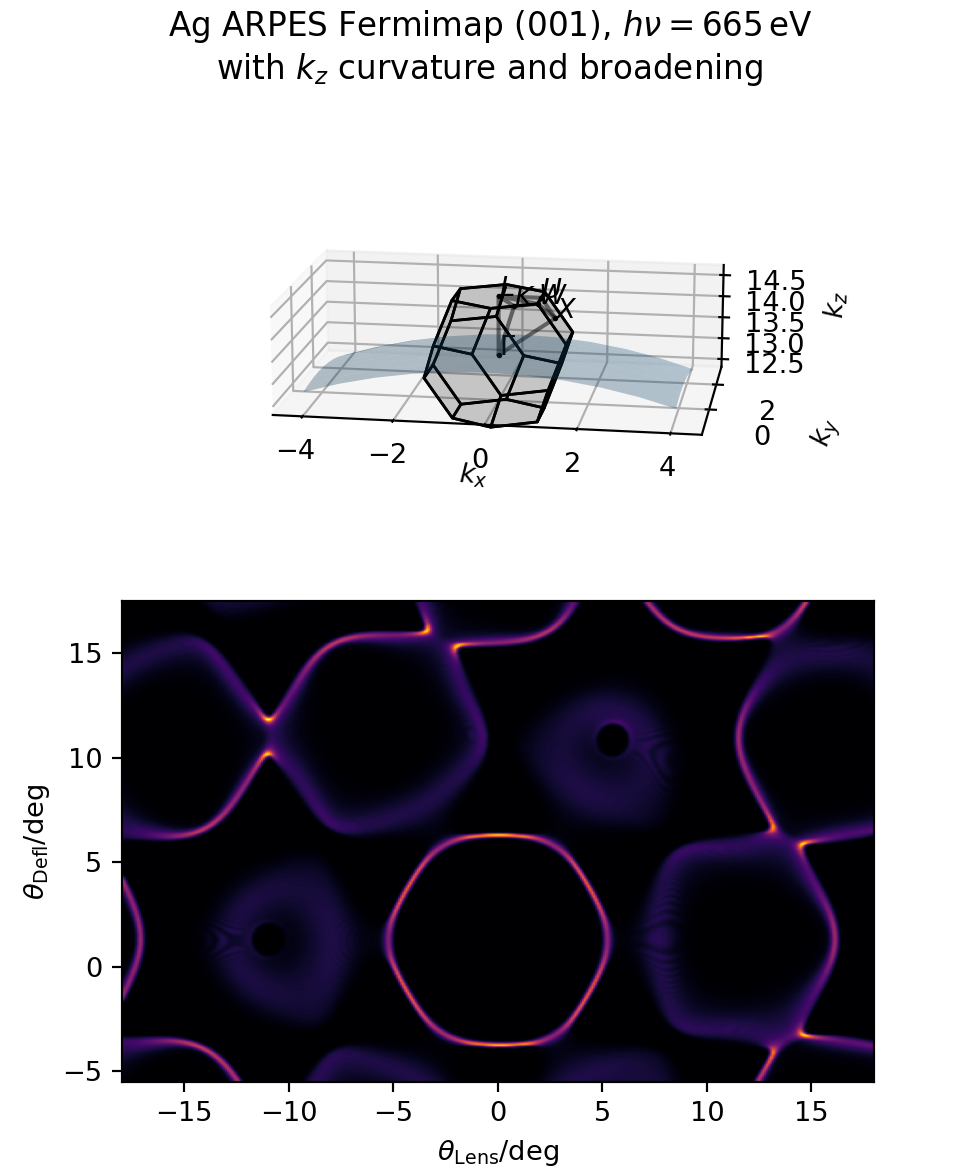
ARPES Fermi surface map simulation¶
/home/docs/checkouts/readthedocs.org/user_builds/fplore/envs/latest/lib/python3.8/site-packages/pymatgen/symmetry/kpath.py:178: UserWarning: The input structure does not match the expected standard primitive! The path can be incorrect. Use at your own risk.
warn(
Getting interpolator...
Got interpolator. Doing interpolation...
import numpy as np
from scipy.stats import norm
from fplore import FPLORun
from fplore.util import rot_v1_v2
from fplore.plot import plot_bz
from fplore.arpes import k_arpes, TPS45A1
import matplotlib.pyplot as plt
kz_broadening = True
kz_curvature = True
e_broadening = True
kxky_broadening = True
run = FPLORun("../example_data/Ag")
analyzer_direction = run.high_symm_kpoints['L'] # 111
lens_axis = run.high_symm_kpoints['X'] # 100
# vector rejection of normal to get perpendicular component to surface normal:
lens_axis = lens_axis - (lens_axis @ analyzer_direction) / (analyzer_direction @ analyzer_direction) * analyzer_direction
R = rot_v1_v2(analyzer_direction, [0, 0, 1])
R = rot_v1_v2(R @ lens_axis, [1, 0, 0]) @ R
# usage:
# for multiplying from the left R @ calc coords(3) -> msmt coords(3)
# R.T @ msmt coords(3) -> calc coords(3)
# for multiplying from the right: calc coords(...,3) @ R.T -> msmt coords(...,3)
# msmt coords(...,3) @ R -> calc coords(...,3)
thetacenter, thetawidth = 0, 36
mintheta, maxtheta = thetacenter - thetawidth / 2, thetacenter + thetawidth / 2
theta = np.linspace(np.deg2rad(mintheta), np.deg2rad(maxtheta), 401)
theta2center, theta2width = 6, 23 # 1.3, 12
mintheta2, maxtheta2 = theta2center - theta2width / 2, theta2center + theta2width / 2
theta2 = np.linspace(np.deg2rad(mintheta2), np.deg2rad(maxtheta2), 400)
extent = (mintheta, maxtheta, mintheta2, maxtheta2)
theta, theta2 = np.meshgrid(theta, theta2, indexing='ij')
v0 = 10
workfunc_analyzer = 3.8
e_photon = 665
if kz_broadening:
imfp = 10 # angstroms, see https://doi.org/10.1384/jsa.9.285
Delta_kz = 1 / imfp # standard deviation in reciprocal angstroms (∆o)² = <o²> - <o>²
surface_normal = R @ analyzer_direction # perfectly flat surface perpendicular to analyzer direction
n_kz = 47 # 13
kz_deltas = np.linspace(-3 * Delta_kz, 3 * Delta_kz, n_kz) # shape (N,)
kz_weights_pdf = norm.cdf(kz_deltas, scale=Delta_kz)
kz_limits = np.hstack([[-np.inf], (kz_deltas[1:] + kz_deltas[:-1]) / 2, [np.inf]])
kz_weights = np.array([norm.cdf(u, scale=Delta_kz) - norm.cdf(l, scale=Delta_kz)
for l, u in zip(kz_limits, kz_limits[1:])])
weights = kz_weights.reshape((1, 1, n_kz, 1))
kz_deltas = kz_deltas.reshape((1, 1, n_kz, 1)) * surface_normal.reshape(1, 1, 1, 3) # to shape (1, 1, N, 3)
else:
kz_deltas = 0
weights = 1
k_p_mean = k_arpes(theta=theta, e_photon=e_photon, phi_det=workfunc_analyzer, v0=v0, theta2=theta2, geometry=TPS45A1)
if not kz_curvature:
k_p_mean[:, :, 2] = k_p_mean[:, :, 2].max()
k_p = k_p_mean[:, :, np.newaxis] + kz_deltas # add kz broadening sample points in 3rd dimension
k = k_p @ R # aligned -> calculation (since R == R⁻¹.T)
k_1bz = run.backfold_k(k)
fig = plt.figure(figsize=(5, 6), constrained_layout=True, dpi=196)
#fig.subplots_adjust(wspace=0.6, hspace=0.6)
ax_bz = fig.add_subplot(2, 1, 1, projection='3d')
ax_bz.plot_surface(*(k_p_mean[::40, ::40]).T,
alpha=0.3) # don't draw all the points # - [0, 0, 8] @ run.primitive_lattice.reciprocal_lattice.matrix @ R
midpoint = k.mean(axis=(0, 1, 2))
offset = midpoint @ run.primitive_lattice.reciprocal_lattice.inv_matrix
offset = np.rint(offset)
plot_bz(run, ax_bz, k_points=False, vectors=None, offset=offset, rot=R.T)
ax_bz.view_init(elev=15., azim=-81.)
ax = fig.add_subplot(2, 1, 2)
ax.set_xlabel(r'$\theta_\mathrm{Lens} / \mathrm{deg}$', )
ax.set_ylabel(r'$\theta_\mathrm{Defl} / \mathrm{deg}$')
level_indices = run.band.bands_within(0.2) # (*energy_window)
print('Getting interpolator...')
ip = run.band.get_interpolator(bands=level_indices)
print('Got interpolator. Doing interpolation...')
data = ip(k_1bz)
if not e_broadening:
scale = 0.01 # FWHM, eV
else:
scale = 0.1 # FWHM, eV
scale = scale / 2.355 # FWHM -> std
im = run.band.smooth_overlap(data, e=0, scale=scale, axis=2, weights=weights)
if kxky_broadening:
from scipy.ndimage import gaussian_filter
im = gaussian_filter(im, (1, 1))
ax.imshow(im, extent=extent, origin='lower', aspect='equal', cmap='inferno')
plt.suptitle(f'Ag ARPES Fermimap (001), $h\\nu = {e_photon}\\,\\mathrm{{eV}}$\nwith $k_z$ curvature and broadening')
ax_bz.set_box_aspect([ub - lb for lb, ub in (getattr(ax_bz, f'get_{a}lim')() for a in 'xyz')])
plt.show()
Total running time of the script: (0 minutes 10.169 seconds)